Nat Commun: Using DNA-Cas9 to map DNA mutations
Release date: 2017-11-28
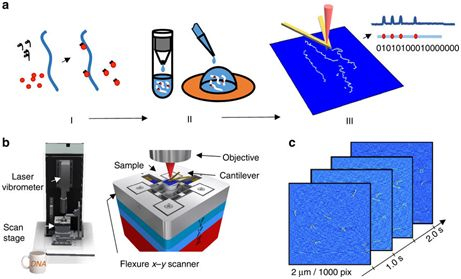
Image courtesy of Nature Communications, doi: 10.1038/s41467-017-01891-9.
In a new study, Dr. Jason Reed and colleagues from the Commonwealth University of Virginia developed a new nanomapping technique that could lead to a revolution in the diagnosis and discovery of pathogenic genetic mutations. This new technology combines high-speed atomic force microscopy (AFM) with a CRISPR-based chemical barcode technology to accurately map DNA as much as DNA sequencing, while processing large fragment genomes faster. More importantly, this technology can be powered by components found in ordinary DNA players. The results of the study were published online November 21, 2017 in the journal Nature Communications, entitled "DNA nanomapping using CRISPR-Cas9 as a programmable nanoparticle".
The human genome consists of billions of DNA base pairs. Once loose, it is nearly 6 feet long. When cells divide, they must copy their DNA for new cells. However, sometimes multiple fragments of this DNA are erroneously copied or linked together at the wrong site, resulting in mutations in genes that cause diseases such as cancer. DNA sequencing is as precise as it can analyze a single base pair of DNA. But in order to analyze large genomes to discover genetic mutations, technicians must identify millions of tiny sequences and then stitch them together using computer software. In contrast, biomedical imaging techniques such as fluorescence in situ hybridization (FISH) are only capable of analyzing DNA at resolutions of tens of thousands of base pairs.
Reed's new high-speed AFM method maps DNA at tens of base pairs and creates images with resolutions up to one million base pairs. And the number of samples it uses is only a fraction of what is needed for DNA sequencing.
Reed said, "DNA sequencing is a powerful tool, but it still has high costs and has some technical and functional limitations that make it difficult to efficiently and accurately map large fragments of genomes. Our approach Fills the gap between DNA sequencing and other lower resolution physical mapping techniques. It can be used as an independent method and can be used as a complement to DNA sequencing to reduce the genome of small fragments during DNA sequencing. The complexity and errors when stitching together."
The AFM draws a picture of the surface of the material by contacting its microscopic stylus with the surface of the material being studied. However, traditional AFM is too slow for medical applications, so it is primarily used by materials science engineers.
Reed said, "Our equipment works the same way as AFM, but lets the sample pass through the microneedle at a greater rate and uses optical instruments to detect the interaction between the microneedle and the molecules on the surface of the material. It can achieve the same observation details as traditional AFM, but can process materials a thousand times faster than the latter. High-speed AFM is very suitable for some medical applications because it can process materials quickly and provide better imaging methods than similar ones. Hundreds of resolutions."
Increasing the speed of AFM is only one obstacle that Reed and his colleagues must overcome. In order to truly identify genetic mutations in DNA, they must develop a method of placing markers or labels on the surface of DNA molecules so that they can recognize patterns and irregularities in these molecules. They also used CRISPR technology to develop a clever chemical barcode method.
Recently, due to advances in gene editing, the CRISPR/Cas9 system has become the headline of many news reports. The scientists used the guide RNA to "program" the enzyme Cas9 to cleave the DNA at the exact site, and then the cell was able to repair the DNA damage itself. The Reed team changed the chemical reaction conditions of this enzyme so that it only binds to DNA, but does not cut it.
Reed said, "Since Cas9 is a protein that is physically larger than DNA molecules, it is perfect for this barcode application. We were surprised to find that its efficiency in binding to DNA molecules is close to 90%. Because observing Cas9 is relatively easy, you can find mutations in the DNA pattern."
To confirm the validity of this technique, these researchers mapped the gene translocations present in lymph node biopsies from patients with lymphoma. These gene translocations are particularly prevalent in blood cancers such as lymphoma, but are also present in other cancers.
Although this technology has many potential uses, Reed and his team are focusing on medical applications. They are currently developing software based on existing algorithms to be able to analyze patterns in DNA fragments of one million base pairs and above. Once development is complete, it is not difficult to imagine that in a pathology laboratory, this shoebox-sized instrument will help diagnose and treat diseases associated with genetic mutations.
References Andrey Mikheikin, Anita Olsen, Kevin Leslie et al. DNA nanomapping using CRISPR-Cas9 as a programmable nanoparticle. Nature Communications, Published online: 21 November 2017, doi:10.1038/s41467-017-01891-9
Source: Bio Valley
Etheric Compound,Dimethyl Sulfoxidum,Dimethyl Sulphoxide,Dimethyl Sulfoxide
Wuxi Yangshan Biochemical Co.,Ltd. , https://www.yangshanchem.com